Ressources and databases
biologievegetale.be
Ce site web est un outil pédagogique regroupant plusieurs modules dédicacés à la biologie végétale, depuis la systématique jusqu’à l’écologie en passant par la physiologie. Vous pouvez donc vous initier et vous perfectionner dans ces matières, en particulier en réalisant les quizz et autres activités. Ce site est mis à jour régulièrement car il est utilisé en partie dans le cadre d’enseignements à l’UCLouvain.

Botalgorithme
BotAlgorithme is a project that aims at making plant computational models accessible to an audience as large as possible. We are convinced that plant models are unique tools for teaching complex plant processes. Currently, our preferred process is the regulation of the root water intake.
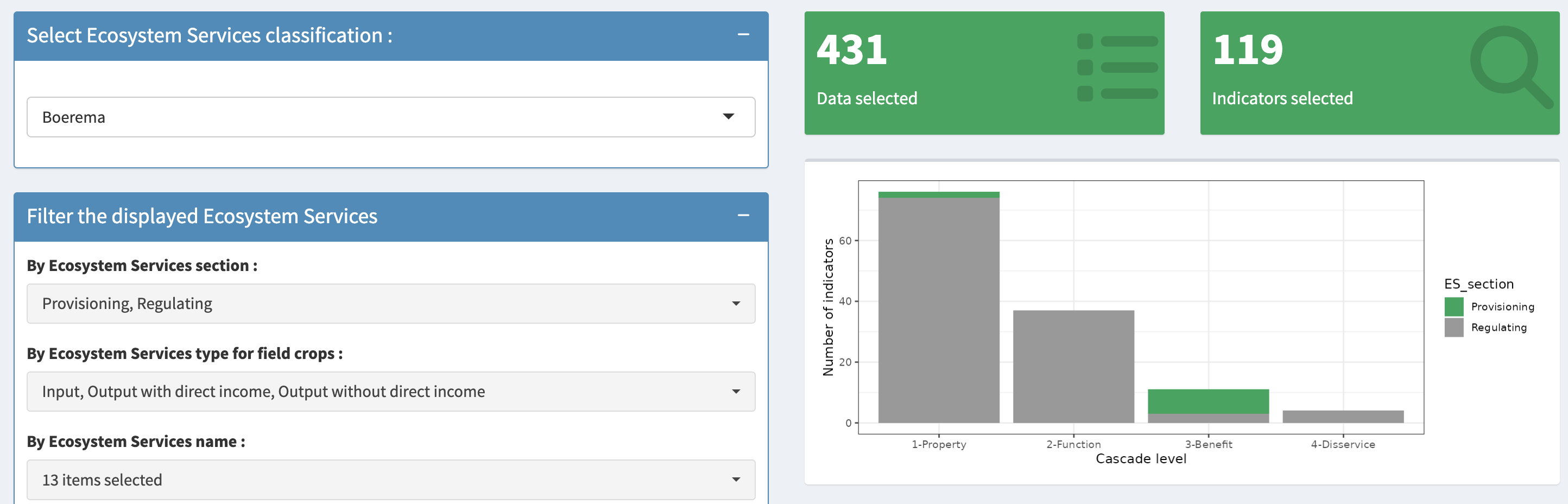
Field-based indicators for biophysical assessment of Ecosystem Services in crop fields
This website presents a list of indicators that can be used for quantifying provisioning and regulation ecosystem services (ES) in crop fields via empirical measure methods. It aims to support researchers and practitioners in selecting context-specific indicators that enhance site-specific knowledge about ES in crop fields, for the development of sustainable cropping systems.
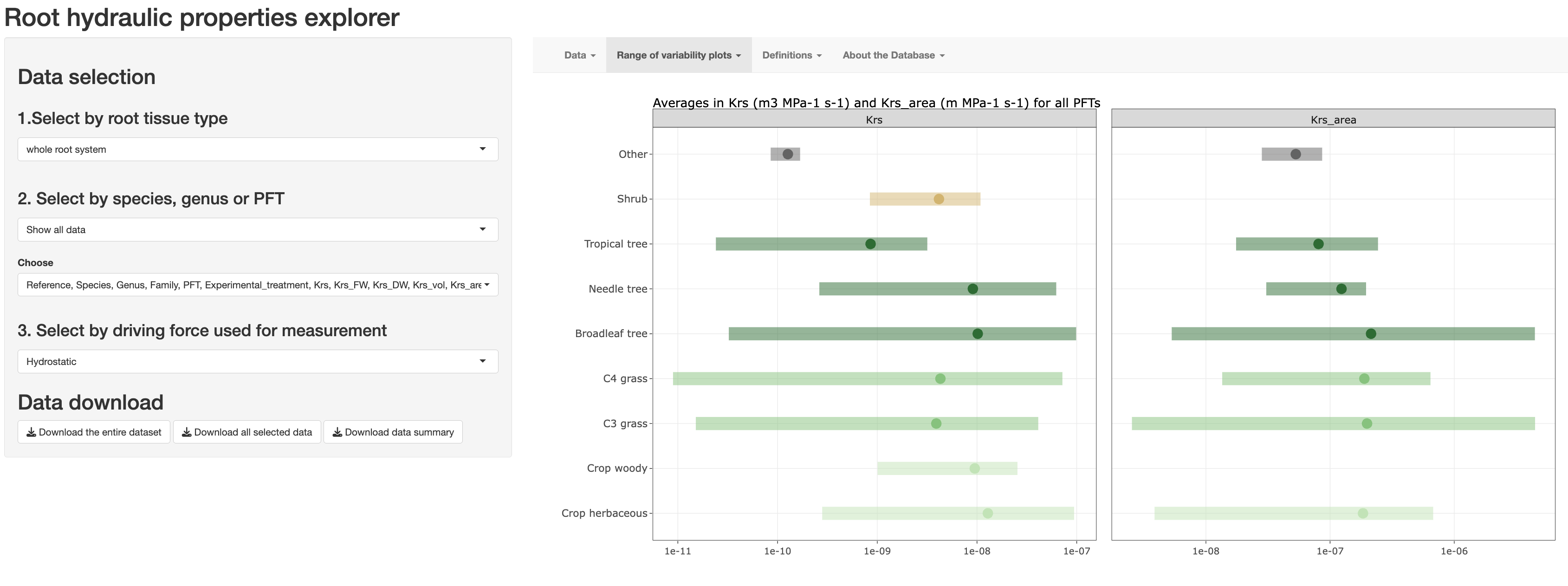
Root Hydraulic Database
We compiled an online database that corresponds to a systematic review of scientific articles in which root hydraulic properties were determined empirically. The relevant data presented in the publications was digitilized manually and can be easily accesed through this application.
quantitative-plant.org
The Quantitative Plant website is an online, manually curated, database referencing more than 180 plant image analysis software solutions, 30 plant image datasets and 100 plant computational models. The website presents each ressource in a uniform and concise manner enabling users to identify the available solutions for their experimental needs. The website also enables user feedback, evaluations and new software submissions. The aim of such a toolbox is to help users to find solutions, and to provide developers a way to exchange and communicate about their work.
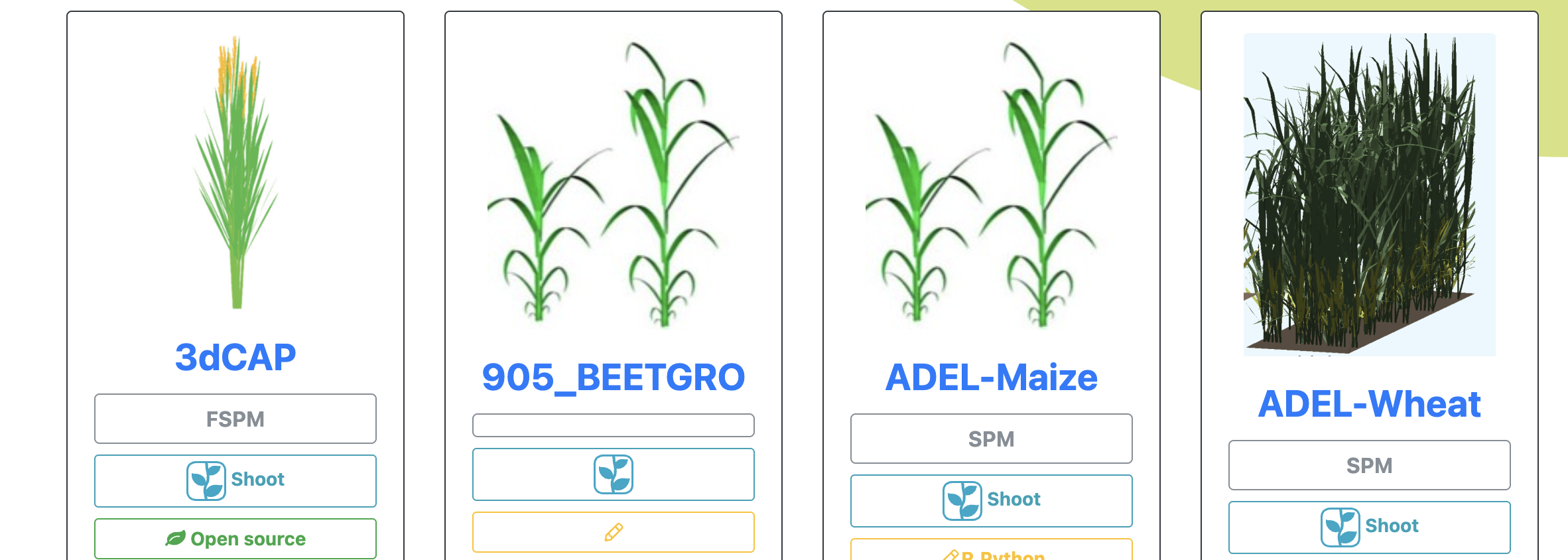
Computational models
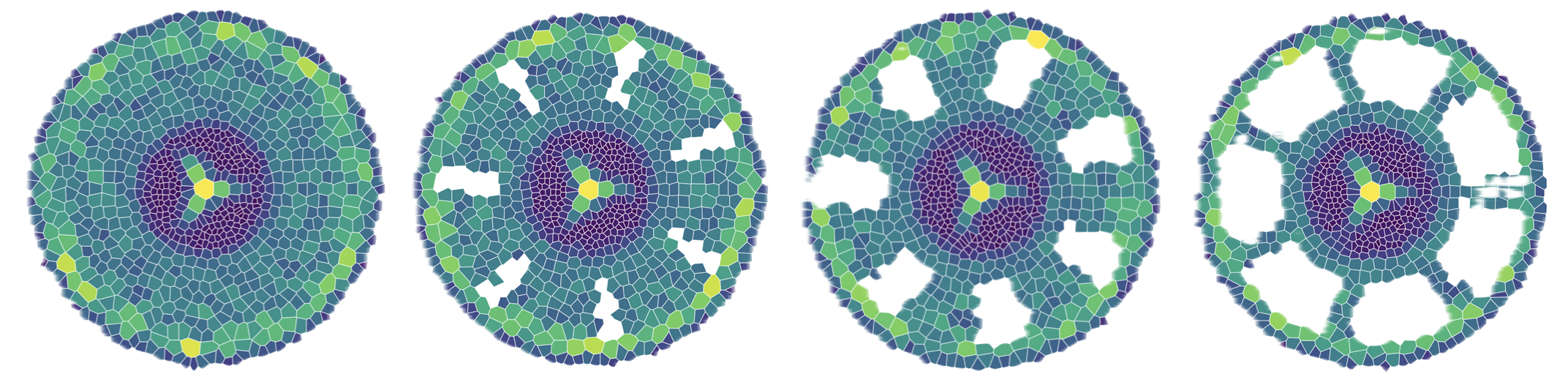
GRANAR
GRANAR is a generator of complete root cross-section network in R. Upon a small set of anatomical features the GRANAR model is able to reconstruct a generic root cell network for mono and dicotyledon. Among the few parameters you can modify, there is the aerenchyma proportion, the number and size of the xylem vessels, but also the number of cell layers and the mean size of each cell type.
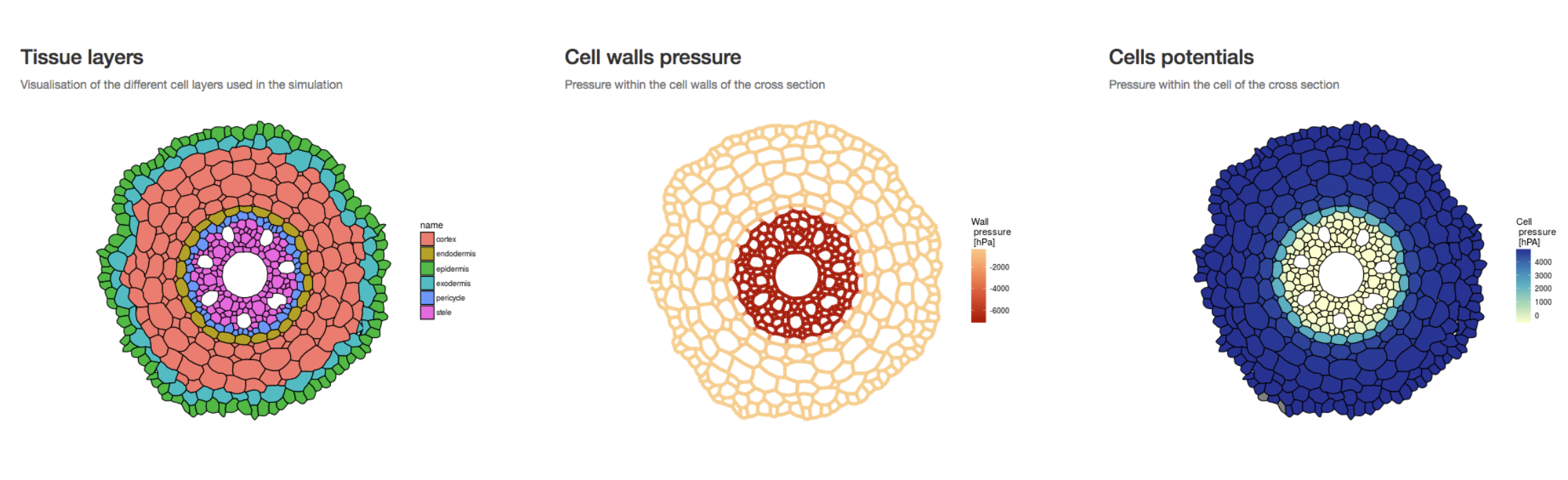
MECHA
MECHA is an explicit cross-section model of the root hydraulic anatomy which connects hydraulic concepts across scales. The model computes horizontal water flow at the level of individual cells, quantifies the contribution of water composite pathways, and predicts root radial permeability (kr), using detailed anatomical descriptions and experimental data on the permeability of cell walls (kw), membranes (Lp) and the conductance of individual plasmodesmata (KPD).
CRootBox / CPlantBox
The focus of CRootBox is the simulation of different types of root architecture, and to provide a generic interface for coupling with soil/environmental models, e.g., in order to determine the impact of specific root architectures on function. CRootBox is currently being extended into a full plant model CPlantBox

Phenotyping tools
RootPhair
In the RootPhair aeroponics platform, roots growing unimpeded can be accessed non-invasively. With backlight illumination, sharp images are produced on 1000 plants with a 2-hr time resolution. The short displacement of roots in one time lapse makes it possible to track the progression of root tips during more than two weeks.

SmartRoot
SmartRoot is a semi-automated image analysis software which streamlines the quantification of root growth and architecture for complex root systems. The software combines a vectorial representation of root objects with a powerful tracing algorithm which accommodates to a wide range of image source and quality. The software supports a sampling-based analysis of root system images, in which detailed information is collected on a limited number of roots selected by the user according to specific research requirements.

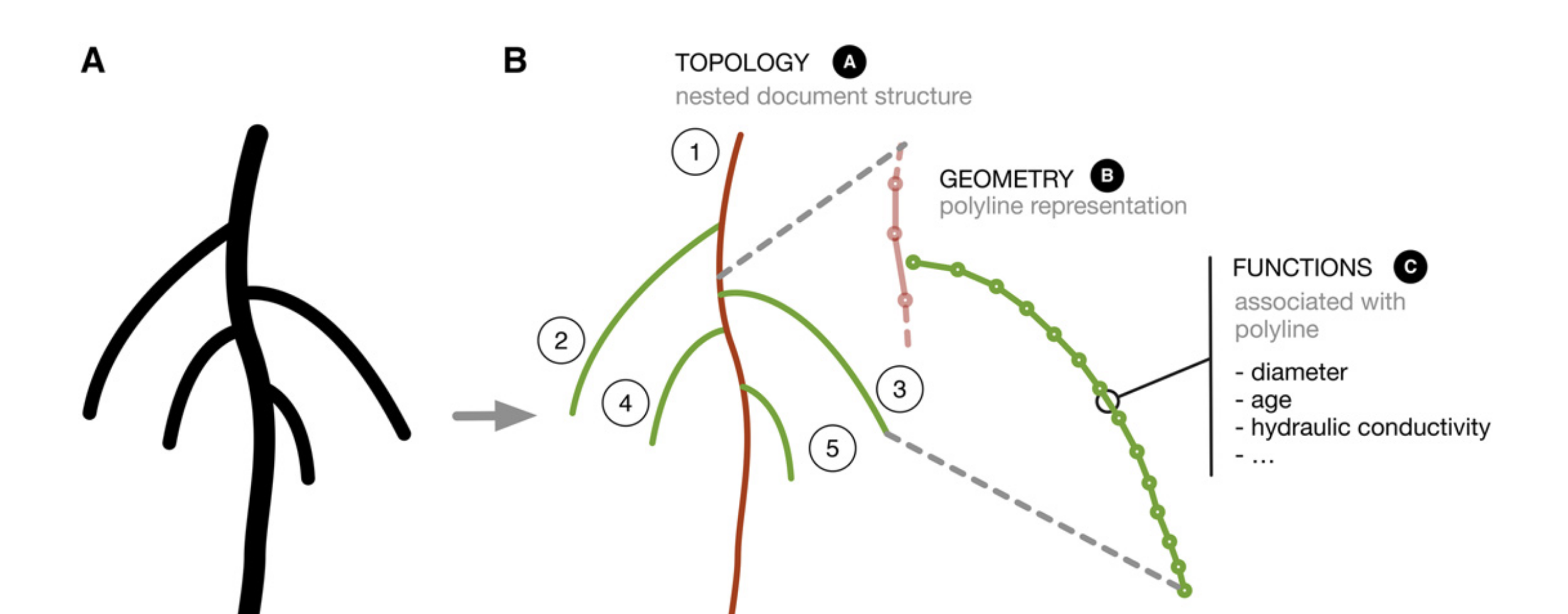
Root System Markup Language | RSML
Together with researchers from INRIA, CPIB, Juelich, Vienna and Louvain, we created the Root System Markup Language (RSML) that has been designed to alleviate two major bottlenecks: (i) to enable portability of root architecture data between different software tools in an easy and interoperable manner allowing seamless collaborative work, and (ii) to provide a standard format upon which to base central repositories which will soon arise following the expanding worldwide root phenotyping effort.